Optical Manipulation and Imaging
The Molecular Mechanobiology team develops novel single molecule manipulation and imaging tools for molecular biology. Manipulation of single molecules is realized using high-resolution optical tweezers, which allow probing sub-nanometer conformational changes of proteins or nuclei acids with sub-millisecond time resolution. Imaging and 3D localization of single molecules with nanometer accuracy is performed through fluorescent probes and advanced microscopy approaches in vitro as well as in living cells. We apply these techniques to the study of molecular motors, gene expression, and mechanical regulation of biological systems.

We developed an ultrafast force-clamp spectroscopy technique that uses a dual trap configuration to apply constant loads between a single intermittently interacting biological polymer and a binding protein [1]. Our system has a delay of only ~10 μs between formation of the molecular bond and application of the force, and can detect interactions as short as 100 μs. The force-clamp configuration in which our assay operates allows direct measurements of the load dependence of the lifetimes of single molecular bonds. Moreover, conformational changes of single proteins and molecular motors can be recorded with sub-nanometer accuracy and a temporal resolution in the tens of microseconds. We applied our technique to the study of molecular motors, using myosin II from fast skeletal muscle, and to protein-DNA interaction, for the lactose repressor (LacI) interaction with DNA.
[1] Capitanio M., Canepari M., Maffei M., Beneventi D., Monico C., Vanzi F., Bottinelli R., and F. S. Pavone, Ultrafast force-clamp spectroscopy of single molecules reveals load dependence of myosin working stroke, Nature Methods 9, 1013–1019 (2012) full text, cover page, author's file
[2] Capitanio M. and F. S. Pavone, Interrogating biology with force: single molecule high resolution, Biophysical Journal, Vol. 105, Issue 6, pp. 1293-1303, 2013, September 17. full text
Ultra-fast optical tweezers
3D localization of single molecules

We developed a novel method for three-dimensional localization of single nano-emitters based on automatic recognition of out-of-focus diffraction patterns. Our technique can be applied to track the movements of single molecules in living cells using a conventional epifluorescence microscope. Three-dimensional localization of fluorescent nanobeads is obtained over 4 microns depth with accuracy below 2 nm in vitro. We also establish three-dimensional tracking of Quantum Dots, overcoming their anisotropic emission, by adopting a ligation strategy that allows rotational freedom of the emitter combined with proper pattern recognition. We localize commercially available Quantum Dots in living cells with accuracy better than 7 nm over 2 microns depth.
Important localization errors can occur in out-of-focus imaging when improperly calibrated and we give indications to avoid them. Finally, we share a Matlab script that allows readily application of our technique by other laboratories.
[3] Gardini, L., Capitanio, M., Pavone F.S., “3D tracking of single nanoparticles and quantum dots in living cells by out-of-focus imaging with diffraction pattern recognition”, Scientific Reports 5, 16088; doi: 10.1038/srep16088 (2015).
Combined manipulation and imaging
Single molecule (SM) techniques have greatly developed over the past thirty years to respond to the need of overcoming some of the limitations of traditional, bulk solution measurements. The manipulation of single biological molecules has created the opportunity to measure mechanical properties of biopolymers and control the mechanical parameters of protein-protein and protein-DNA interactions. SM fluorescence detection, on the other hand, represents an incredibly versatile tool for studying protein activity in vitro and in vivo, leading to the possibility of localizing and tracking single molecules with nanometer precision.
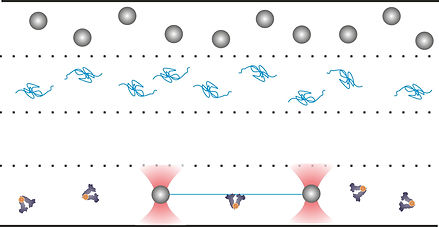
Through fitting of the instrument point-spread-function to the SM image, in fact, one can accomplish localization with a precision depending mainly on signal-to-noise ratio (SNR) and reaching a limit of about one nanometer. These methodologies find powerful applications in the study of the dynamics of motor proteins, as well as of the diffusion processes underlying target search in DNA-binding proteins. The capability of determining diffusion constants as a function of the DNA sequence, residence time on the target and accurately measuring the DNA length explored during one-dimensional diffusion events, represent a powerful tool for the study of protein-DNA interaction dynamics and for the investigation of the mechanisms of specific target search.
Recently, the combination of these two techniques has produced a new generation of experimental setups enabling the simultaneous manipulation of a biological substrate (for example an actin filament or a DNA molecule) and detection/localization of an interacting partner enzyme (for example myosin or a DNA-binding protein). The advantages of these techniques mainly rest on the possibility of exerting mechanical control over the trapped polymer, thus enabling the study of interaction dynamics versus forces or torques. Also, the methodology allows measuring biochemical reactions far from the surface, avoiding one of the main limitations of classic SM methods, i.e., the need for immobilization of the molecules under study on a surface (glass slide or microspheres).
The combination of two single molecules techniques requires overcoming several technical difficulties, mainly arising from the requirements of mechanical stability and adequate SNR (especially when requiring localization with nm precision). Particularly, when coupling SM fluorescence detection with optical tweezers, the reduction of noise and photobleaching from the trapping infrared lasers and the control of biochemical buffers for assembly of the biological complexes and performance of the experimental measurements are of paramount importance. We developed methods for performing successful measurements in a dual trapping/SM Fluorescence localization setup. The methodology has been applied to the study of lactose repressor protein (LacI), fluorescently labeled with Atto532, and detected as it binds to a DNA molecule (trapped between two optical tweezers) containing specific LacI binding sequences (i.e., operators). The method is effective in detecting binding of LacI to DNA and diffusion along its contour in the target search process. The method is applicable to any combination of DNA sequence and DNA-binding protein, as well as to other systems (microtubules or actin filaments and the motor proteins interacting with them).
[3] Monico C., Belcastro G., Vanzi F., Pavone F. S., and M. Capitanio. Combining Single-molecule Manipulation and Imaging for the Study of Protein-DNA Interactions. J. Vis. Exp. (90), e51446, doi:10.3791/51446 (2014). (full text)
[4] Monico C., Capitanio M., Belcastro G., Vanzi F., and F. S. Pavone, Optical methods to study protein-DNA interactions in vitro and in living cells at the single-molecule level, International Journal of Molecular Sciences, Vol. 14, n. 2, pp. 3961-3992, 2013, February 18. (full text)
[5] Capitanio M., Maggi D., Vanzi F., and F. S. Pavone, FIONA in the trap: the advantages of combining optical tweezers and fluorescence, J. Opt. A: Pure Appl. Opt., vol. 9, pp. S157-S163, DOI:10.1088/1464-4258/9/8/S07, Cit. 7, 2007 August 1. (full text)
Single Molecule Localization Microscopy
Super-resolution fluorescence microscopy techniques have become increasingly popular over the last decade, owing to the fact that they allow investigators to resolve details orders of magnitude smaller than the optical resolution limit, without having to resort to methods such as electron or scanning probe microscopy techniques. Among the many available super-resolution techniques, the highest reported imaging resolutions have been achieved by localization-based methods such as STochastic Optical Reconstruction Microscopy (STORM) and PhotoActivated Localization Microscopy (PALM), which rely on chemical control over fluorophore photophysics in order to resolve the spatial position of individual fluorescent molecules with nanometer-level accuracy. In our lab we set up novel optical tecniques to achieve improved three-dimensional super-resolution images through a combination of inclined illumination and astigmatism to acquire volumetric super-resolved images of thick samples. We apply 3D STORM microscopy to study the structure of the actin cortex of eukaryotic cells and how it is regulated by the extracellullar mechanical environment. Moreover, PALM microscopy is applied to the sytudy of membrane proteins distribution on planktonic bacteria and bacteria biofilms to investigate the molecular mechanisms at the basis of antibiotic resistance. Lastly, we adopt a combination of single.molecule three-dimensional tracking and super-resolution imaging to study the dynamics of interaction of ERK5, a kinase involved in melanoma growth, with the nucleus.